- In-Fusion® HD Multiple-Insert Cloning Protocol-At-A-Glance (121416) takarabio.com Takara Bio USA, Inc. Page 1 of 7 Please read the In-Fusion HD Cloning Kit User Manual before using this Protocol-At-A-Glance.
- TOPO® TA Cloning® Kit. For Research Use Only. Not for use in diagnostic procedures. 2 Information in this document is subject to change without notice. Supplied with Cat. K4510-02 and K4500-02 refer to the manual supplied with the miniprep kit.
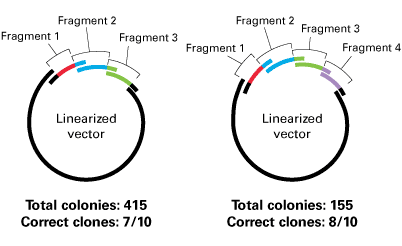
BD In-Fusion™ Method efficiently clones a range of insert sizes. BD In-Fusion Cloning was performed using 100 ng linear vector and 50 ng of each of the PCR products indicated. 1 µl of each 20 µl reaction was then transformed into BD Fusion-Blue™ competent cells.
Description
Intact Genomics propriety ig-Fusion™ cloning technology is a simple, rapid and highly efficient cloning kit which allows to directly clone any PCR product(s) to any linearized expression vector at any site. The PCR fragments can be generated by Intact Genomics high fidelity Pfu DNA polymerase or other high fidelity DNA polymerases, with primers having 15 to 18 bases of homology at their linear ends to where the product need to fuse. The linearized vector can be generated by PCR or restriction enzymes. The kit is so robust that multiple DNA fragments can be assembled simultaneously and cloned into one construct in a single reaction step within short times (usually 10-30 min) with more than 95% cloning efficiency.
Applications
- Clone any insert at any site within any vector
- Restriction enzyme and phosphatase free system
- Joining multiple large fragments at once
- Precise insertion at a desired orientation
- Rapid and high efficiency with > 95% positive clones
Product Includes
- 5x ig-Fusion™ enzyme premix
- 2x PCR premix
- High efficiency competent cells
Storage Temperature
5x ig-Fusion™ enzyme premix: -20 ºC
2x PCR premix: -20 ºC
High efficiency chemical competent cells: -80 ºC4111 4115
In Fusion Cloning Tools
Abstract
We present a method that allows simultaneous fusion and cloning of multiple PCR products in a rapid and efficient manner. The procedure is based on the use of PCR primers that contain a single deoxyuridine residue near their 5′ end. Treatment of the PCR products with a commercial deoxyuridine-excision reagent generates long 3′ overhangs designed to specifically complement each other. The combination of this principle with the improved USER cloning technique provides a simple, fast and very efficient method to simultaneously fuse and clone multiple PCR fragments into a vector of interest. Around 90% positive clones were obtained when three different PCR products were fused and cloned into a USER-compatible vector in a simple procedure that, apart from the single PCR amplification step and the bacterial transformation, took approximately one hour. We expect this method to replace overlapping PCR and the use of type IIS restriction enzymes in many of their applications.
INTRODUCTION
Seamless PCR product fusion is the process by which two or more PCR products are joined precisely so that no undesired base pairs are added at the junction sites. This is the ideal situation for the countless cases in which the fusion of PCR products is required (). The current method of choice when facing the challenge of seamlessly assembling several DNA fragments is overlapping PCR, also called fusion PCR or overlap extension PCR (2–4). This method was introduced shortly after the invention of PCR in the mid-1980s and continues to be the preferred method. The technique is versatile and efficient, but requires several consecutive PCR reactions and, therefore, carries an inherent high risk of PCR-introduced errors. Furthermore, primer design and PCR conditions usually require optimization when multiple DNA fragments of considerable size are to be assembled ().
Uracil excision-based cloning was conceived in the early 1990s as an alternative to restriction digest and ligation-based cloning (6–8). The original method relies on the use of PCR primers that contain an ∼12-nt 5′ tail in which at least four deoxyuridines (Us) have been placed instead of deoxythymidines (Ts). The PCR product is treated with uracil DNA glycosidase (UDG), which excises uracil bases selectively while leaving the phosphodiester backbone intact (). The abasic sites destabilize base pairing, which in praxis results in 3′ overhangs that can anneal to similarly produced complementary 3′ overhangs in a PCR-amplified vector (). Alternatively, suitable 3′ overhangs in a vector can be produced by ligation of a short U-containing DNA sequence to the ends of a linearized vector and further treatment with UDG (). The treated PCR product and vector are able to form a stable circular hybridization product that can be transformed into Escherichia coli without prior ligation (,).
Although early uracil excision-based cloning proved to be an efficient and versatile technique (up to 1 × 107 transformants ) (), several useful modifications have been implemented. Watson and Bennett included a second enzyme in the treatment of PCR products (). T4 endonuclease V is capable of breaking the DNA phosphodiester backbone at the 3′ side of an abasic site (11). Therefore, the sequential activity of UDG and T4 endonuclease V on PCR products amplified with primers containing a single U residue generates 3′ overhangs that can be used efficiently for cloning (). The commercial USER™ Friendly Cloning Kit (New England Biolabs, launched in 2003) makes use of a similar double enzymatic treatment. In this cloning technique, the PCR product is treated with USER™ enzyme mix, a mixture of UDG and DNA glycosylase-lyase endo VIII (the exact proportions are not disclosed by the company). The latter enzyme can break the phosphodiester backbone at the 3′ and 5′ sites of an abasic site so that base-free deoxyribose is released (,). This makes USER™ enzyme mix a deoxyuridine-excision reagent. Furthermore, in the USER™ Friendly Cloning Kit, the supplied vector has been prepared by inserting a small cassette into the multiple cloning site, and subsequently treating the vector with two restriction endonucleases: a linearizing one and a nicking one (http://www.neb.com/nebecomm/ManualFiles/manualE5500.pdf).
Despite the various modifications to the original method, uracil excision-based cloning has remained largely unused (), most probably because of incompatibility with proof-reading DNA polymerases, which stall at deoxyuridines present in DNA templates (,). Recently, Nour-Eldin et al. improved the technique by identifying PfuCx as a compatible proof-reading DNA polymerase and by developing an improved vector design strategy (). We will refer to this technique as ‘improved USER cloning technique’.
In this article, we describe the assembly of an open reading frame composed of three different DNA fragments (ORF1) by means of a deoxyuridine excision-based method that combines the fusion of multiple PCR products with the cloning of the fusion product into a vector in a single step, without ligation. The advantages of ‘USER fusion’ are discussed in comparison with other fusion techniques.
MATERIALS AND METHODS
PCR amplification
Three PCR amplifications were carried out in parallel using plasmid templates containing full-length cDNAs of genes X1 (At2g30860, 648 bp), X2 (At4g31500, 1500 bp) and X3 (At5g05260, 1572 bp), and primer pairs X1-fwd/X1-rev, X2-fwd/X2-rev and X3-fwd/X3-rev, respectively. The sequence of the oligonucleotide primers, all purchased from Invitrogen, are listed in Table 1. The reactions were carried out using PfuTurbo® CX Hotstart DNA polymerase (Stratagene) according to the manufacturer's instructions. Additionally, 5% DMSO was included in the reaction that amplified X2 cDNA.
Table 1.
Oligonucleotide primers used for the assembly of ORF1 by USER fusion
Target-specific sequences are presented in italics. Restriction sites are presented in bold. The identity of each of the underlined sequences is written beneath them. U residues are shaded in black, while the regions shaded in grey correspond to the oligonucleotides that will be displaced after treatment with USER™ enzyme mix.
DNA purification
The PCR products were column-purified using QIAquick PCR purification kit (QIAGEN) according to the manufacturer's instructions. Purified PCR products were eluted with water in one-third of the original volume.
Mixture of purified PCR products and pre-digested vector
An aliquot of the purified PCR products, along with an aliquot of a stock of PacI/Nt.BbvCI pre-digested pCAMBIA330035Su (∼9.5 kb) (), was run on an agarose gel to estimate relative concentrations. The four components were mixed in approximate 10:10:10:1 molar ratios, where the unit refers to pre-digested vector.
Treatment with USER™ enzyme mix
One microliter of 10× TE buffer [100 mM Tris-HCl, 1 mM EDTA (pH 8.0)] and 1 U of USER™ enzyme mix (New England Biolabs, 1 U/μl) were added to 8 µl of the mixture of purified PCR products and pre-digested vector. The reaction mixture was incubated for 20 min at 37°C, followed by 20 min at 25°C. The approximate amount of pre-digested vector in the reaction mixture was 0.01 pmol.
Transformation
The entire USER-treated reaction mixture (10 µl) was used to transform 50 µl of chemically competent E. coli cells by heat shock.
Plasmid preparations and restriction analysis
Plasmid preparations were made from 4 ml overnight cultures using GenElute™ Plasmid Miniprep Kit (Sigma-Aldrich) according to the manufacturer's instructions. The purified plasmids were eluted in 50 μl of water. Two microliters of each plasmid miniprep were cut for 2 h at 37°C with 10 U of SpeI (New England Biolabs) in 20 μl reactions that contained 1×NEB Buffer 2 [50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol (pH 7.9)] and 100 μg/ml BSA.
RESULTS
ORF1 is composed of the coding regions of genes X1, X2 and X3, linked by two different ∼60-bp linkers, L1 (between X1 and X2) and L2 (between X2 and X3) (Figure 1A). The entire ORF1 has a length of ∼3.9 kb.
(A) Outline of ORF1. The coding regions of genes X1, X2, and X3 are linked together by two different ∼60-bp linkers, L1 and L2. ORF1 has a length of ∼3.9 kb. (B) Assembly of ORF1. X1, X2 and X3 sequences were amplified using primers that contained a single U (instead of a T) near their 5′ end. The L1 sequence was included as two separate fragments in primers X1-rev and X2-fwd, and the L2 sequence was similarly included in primers X2-rev and X3-fwd so that both ∼60-bp linkers were constructed after the fusion procedure. The PCR products were mixed with a pre-digested USER-compatible vector and treated with the deoxyuridine-excising USER™ enzyme mix. This generated 3′ overhangs that were designed so that the internal ones complemented each other (indicated by arrows), while the outermost ones complemented the overhangs of the pre-digested vector. This design enabled the formation of a stable circular hybridization product that was transformed directly into E. coli without prior ligation.
The fusion and cloning principle used for the assembly of ORF1 is depicted in Figure 1B. The coding regions of X1, X2 and X3 were amplified using primers that contained a single deoxyuridine (instead of a deoxythymidine) near their 5′ end. The L1 sequence was included as two separate fragments in primers X1-rev and X2-fwd, and the L2 sequence was similarly included in primers X2-rev and X3-fwd so that both ∼60-bp linkers were constructed after the fusion procedure. The deoxyuridine residues in these four primers were located 6–8 residues downstream from a 5′ terminal deoxyadenosine. The purified PCR products were mixed with a pre-digested USER-compatible vector and treated with the deoxyuridine-excising USER™ enzyme mix. Ablation of the deoxyuridines from the PCR products resulted in the generation of 7–9 nt-long overhangs. The 7-nt overhang generated with primer X1-rev was designed to complement the 7-nt overhang generated with primer X2-fwd, and a similar design had been made for the 9-nt overhangs generated with primers X2-rev and X3-fwd. The 8-nt overhangs generated with primers X1-fwd and X3-rev complemented the overhangs present in the pre-digested vector. A short incubation at 25°C ensured the formation of a stable circular hybridization product that was ready for transformation into E. coli without prior ligation. This procedure thus allowed simultaneous fusion and insertion of the three PCR products into the pre-digested vector. When using ∼0.01 pmol of pre-digested vector, ∼100 colonies were obtained in two replicates derived from two separate enzymatic treatments.
Primers X1-fwd and X3-rev carried two unique restriction sites each, between the target-specific sequence and the deoxyuridine residue (Table 1). These sites were included for future sub-cloning purposes. We used SpeI sites present in both primers X1-fwd and X3-rev for restriction analysis of plasmids containing a full-length ORF1. The analysis of plasmid preparations from 32 independent colonies showed an ∼3.9-kb fragment in 29 preparations (Figure 2). Two positive plasmids were sequenced. The sequencing results showed that both ORF1 sequences were free of polymerase-introduced errors and that they had been inserted in the correct orientation.
Agarose gel showing a restriction analysis of potential ORF1-containing plasmids. Plasmids from 32 independent colonies were digested using SpeI enzyme and run on an agarose gel. An aliquot of a molecular weight marker was loaded in the outermost lanes and in the middle lane. An ∼3.9-kb band (black arrow), which represents the full-length ORF1, was seen in 29 out of the 32 digestions (∼90%).
DISCUSSION
We describe a method in which multiple PCR products are fused and cloned simultaneously into a vector of interest. The fusion principle makes use of deoxyuridine-containing primers and an enzymatic treatment to generate long overhangs by 5′ partial primer removal. Overhangs of the individual PCR products are designed to complement the appropriate overhangs of other products, thus making the fusion process possible. The combination of this principle with the improved USER cloning technique (), results in a simple, fast and very efficient method to fuse and clone multiple PCR fragments into a vector in a single step.
The fusion principle was originally introduced by Booth et al. () and by Watson and Bennett () (,). However, their methods do not seem to have been applied elsewhere, as evidenced by the lack of citations. Booth et al. (1994) used the original uracil excision-based cloning (several Us per primer and only UDG for the enzymatic treatment), and managed to clone the fusion product without ligation. In two separate experiments, each involving two PCR products, only two out of forty, or six out of ten randomly selected colonies contained the desired insert (). We have shown that, in the USER fusion of three PCR products, 29 out of 32 randomly selected colonies contained the desired insert (∼90%). The higher percentage of positive colonies is explained by the fact that the USER™ enzyme mix contains both UDG and DNA glycosylase-lyase Endo VIII, and is therefore capable of generating overhangs (see the Introduction section). On the other hand, Watson and Bennett did use two different enzymes to generate overhangs in their PCR products (each primer carrying a single U) and they reported very high cloning efficiencies. However, as opposed to USER fusion, the cloning required ligation, since it was made into a SpeI-cut vector (4-nt overhangs) (). When Nour-Eldin et al. reported the improved USER cloning technique, they suggested a sequential insertion of PCR products in order to achieve fusions. This leads to the inclusion of a 13-bp sequence at the joining site (). The novelty in our method lies in the combination of the fusion principle with the improved USER cloning technique. With USER fusion, multiple PCR products can be fused seamlessly and cloned into a vector of interest in a single step, without ligation.
In USER fusion, the primer design is flexible. The only requirement, when examining the sequence of a desired fusion product, is the presence of a deoxythymidine (T) located ∼8 nt downstream from a deoxyadenosine (A) in the proximity of each junction site (Figure 3). The sequence of the forward primers start with the selected A and has a U instead of the selected T. An analog situation occurs with the reverse primers. This design allows the creation of complementary overhangs of ∼8 nt during the fusion process.
Primer design in USER fusion. The blue lines encompass the desired DNA fusion product, in which X and Y are any pair of complementary deoxynucleotides. Only the sequence near a junction site is shown. The only requirement for USER fusion is the presence of a T located ∼8 residues downstream from an A (both residues shaded in black, as well as their complementary residues) in the proximity of the junction site. The sequences of the reverse primer (that will amplify the upstream fragment) and the forward primer (that will amplify the downstream fragment) are shown. In both primers, the selected T is replaced by a U (shaded in red).
We have successfully used overhangs of 7–9 nt, but longer overhangs are also expected to work efficiently. If very long overhangs are required, the replacement of several Ts for Us in the overlapping parts of the primers (and not only the most downstream T) is recommendable, although this increases the cost of the primers. We recommend having different overhang lengths for the different junction sites because it minimizes the risk of incorrect combinations of overhangs annealing together. However, as a single nucleotide difference in the overhangs used for improved USER cloning is able to efficiently confer directionality (), we predict that similar simultaneous fusion and cloning processes would be efficient even when non-complementary overhangs are of the same length.
We have mentioned that, for USER fusion, appropriate A and T residues must be identified in the desired fusion sequence, proximal to the junction site. The actual location of the selected residues in relation to the junction site may, however, vary. Six different cases, which illustrate the flexibility of the design in USER fusion, are presented in Figure 4.
Strategies for USER fusion. The two colored strands in the middle of each figure represent the desired DNA fusion product. The fragments to be joined together are presented in red (left) and in blue (right). In cases (C), (D), (E) and (F), a linker (gray) is desired at the junction site. The arrows represent the primers designed for USER fusion, and their color code indicates the origin of their sequence. (A) The selected A and T residues surround the junction site. (B) Both residues are chosen at one side of the junction site. (C) The A and T residues lie in the linker itself, and the linker sequence is partially included in each primer. (D) Both residues are chosen at one side of the linker, and the entire linker sequence is included in the primer that amplifies the sequence at the other side. (E) The residues are located at each side of the linker, and the entire linker sequence is included in both primers. (F) The residues are located at each side of the linker, but the linker sequence is not included in any of the primers. Two complementary oligonucleotides that carry the linker sequence are used as an intermediate DNA fragment to be fused.
Based on the high efficiency of the assembly of ORF1 (∼100 colonies per 0.01 pmol of pre-digested vector), we believe that three is not the maximum number of PCR products of similar size that can be simultaneously fused and cloned with USER fusion. This maximum number is likely to be dependent on the size of the individual fragments and recipient vector.
In comparison with overlapping PCR, the primers needed for USER fusion are shorter. Typically 15-nt overlaps are needed for overlapping PCR (18), while deoxyuridine-containing primers need only to have overlaps of ∼8 nt. In contrast to USER fusion, overlapping PCR needs secondary PCR amplification steps and therefore carries a high risk of PCR-induced errors. Finally, overlapping PCR yields a full-length product that for many applications still needs to be cloned into a vector. In USER fusion, the full-length product is cloned into the destination vector in the same step.
Type IIS restriction enzymes cleave outside their recognition sequences (e.g. SapI or Esp3I), and are therefore capable of producing overhangs of any given sequence. When the cleavage is designed to occur between the recognition sites and the core of the PCR products, these enzymes are able to mediate seamless fusion of DNA amplicons (,). In comparison to USER fusion, the overhangs created by type IIS endonucleases are shorter (typically 3 or 4 nt long). This creates a need for ligation prior to transformation. Moreover, the PCR products are required not to contain any of the restriction sites that have been attached to their ends. Apart from being a hurdle in primer design, this condition usually introduces the necessity of using different enzymes or enzyme pairs for each of the PCR products to be fused, which in turn decreases the simplicity of the procedure.
It is worth noting that the fusion principle by itself (when not combined with the improved USER cloning) has several potential applications, now that U-containing PCR products can be amplified with a proof-reading polymerase. For example, one could envision its use to quickly construct a gene disruption cassette (5′-flanking sequence + marker + 3′-flanking sequence) that could be used directly or after ligation (but without secondary PCR amplification or cloning) for gene disruption in organisms that require long flanking homology regions for effective integration into the genome. It might also be useful in assembling full-length infectious viral cDNAs. For these two applications, overlapping PCR and the use of type IIS restriction endonucleases have been the preferred strategies (,).
In conclusion, USER fusion is a simple, fast and very efficient method to fuse seamlessly and clone multiple PCR products into a vector of interest. Because of its positive features, we expect USER fusion to replace overlapping PCR and the use of type IIS restriction enzymes in many of their applications.
ACKNOWLEDGEMENTS
This work was supported by the Danish International Developmental Agency (DANIDA, project 91175) and by the Danish National Research Foundation through its support to the Center for Molecular Plant Physiology (PlaCe). Funding to pay the Open Access publication charge was provided by the Danish National Research Foundation.
Conflict of interest statement. It is noted that the subject matter of this publication is included in an international patent application filed by co-inventors FEG, HHN, BAH, et al. on March 5, 2007, under the Patent Cooperation Treaty. This application claims priority to a United Kingdom patent application, number 0610045.7, filed on May 19, 2006, in connection with Plant Biosciences Limited, a United Kingdom-based technology transfer enterprise.